PicoTwist is involved in nanomanipulation and single-molecule
experiments in biology. Historically, single molecule experiments
were among the first nanotechnologies, appearing in the 1990s to
characterize biopolymers and proteins with unprecedented accuracy. The
idea is that instead of analyzing the behavior of a great number of
molecules as usually done in a test tube, you now analyze the behavior
of a single molecule. Thus instead of measuring average behavior you
have direct access to the biopolymer�s activity in real time.
Let us give examples to illustrate this statement.
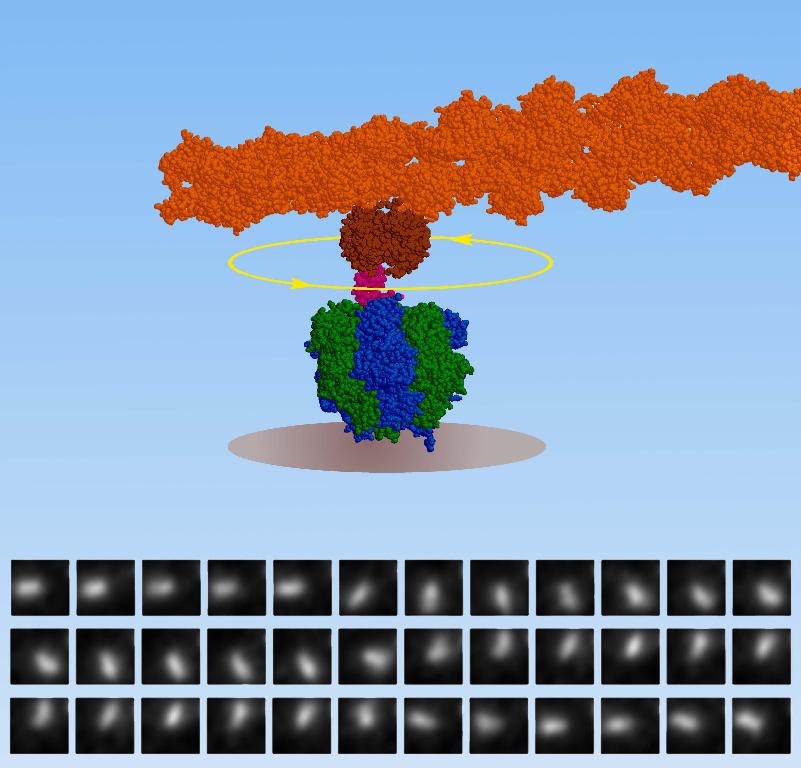
In 1997, the group of
K. Kinosita attached the F1-ATPase unit to a glass coverslip while linking the central part of the enzyme to a rigid, fluorescently labelled protein rod. Upon adding the ATP chemical to the solution, these researchers observed the fluorecent rod spinning furiously. This simple experiment demonstrated that this tiny enzyme, only 10 nm across, is indeed the smallest known
rotary motor spinning in all of our cells
8000 rpm and burning the ATP fuel with nearly 100% efficiency!
Let us dream.
Right now DNA sequencing is achieved by analyzing the length distribution of DNA molecules replicated by millions of DNA-polymerases working on a pool of identical single stranded DNA substrates. This process is very ingenious but relies on the great number of polymerases and DNA molecules which are required to amplify the DNA substrate using the PCR reaction.
The dream of many researchers is to be able to achieve this process on a single DNA substrate. A single DNA polymerase, for instance, is a wonderful machine: it actually reads the sequence of its substrate and places the complementary nucleotide right in place. By fluorescently marking the A nucleotide one may be able to detect each time the
polymerase incorporates it, and we would obtain the position of the A nucleotides along the substrate. Repeating this procedure for each successive nucleotide would provide the full DNA sequence. This dream is not far from becoming reality : using fluorescently labeled nucleotides the groups of
W.W. Webb and
S. Quake have achieved the first steps towards this goal while
S. Block revealed recently (2005 Biophysical meeting) that they were able to detect 3 Angstrom displacement of a single RNA-polymerase as it translocates, base by base, along the DNA.
Picotwist in single molecule experiments.
To achieve such goals requires tracking of a single enzyme working on
a single DNA molecule. This may sound like a real challenge but it is
in fact done every day in research laboratories thanks to single
molecule techniques. Among them, PicoTwist has chosen to develop a
Magnetic Tweezer apparartus. This technique has proven to be powerful
and, unlike other nanomanipulation techniques, extremely stable and
robust. PicoTwist develops and brings to market Magnetic Tweezer
devices which allow one to "dial in" a force and a torque on a
biopolymer such as a DNA molecule, while simultaneously measuring its extension, in
real time and to nanometer resolution. This apparatus is an
ultrastable optical microscope with PC-driven control of tweezing
magnets, optics, and video analysis. This apparatus was built using
the scientific expertise of the Bensimon/Croquette group in Paris.
| |
Bibliography |
| 1 | Direct observation of the rotation of F-1-ATPase
H. Noji, R. Yasuda, M. Yoshida, K. Kinosita,
Nature
(1997) 386-6622 p.299 PubMed CrosRef
|
| 2 | Stepping rotation of F-1-ATPase visualized through angle-resolved single-fluorophore imaging
K. Adachi, R. Yasuda, H. Noji, H. Itoh, Y. Harada, M. Yoshida, K. Kinosita,
Proc. Natl. Acad. Sci. U. S. A.
(2000) 97-13 p.7243 PNAS publications
CrosRef
|
| 3 | Resolution of distinct rotational substeps by submillisecond kinetic analysis of F-1-ATPase
R. Yasuda, H. Noji, M. Yoshida, K. Kinosita, H. Itoh,
Nature
(2001) 410-6831 p.898 PubMed CrosRef
|
| 4 | Single molecule DNA sequence profiling in zero-mode waveguides using gamma-phosphate linked nucleotide analogs
J. Korlach, M. Levene, M. Foquet, SW. Turner, HG. Craighead, WW. Webb,
Biophys. J.
(2003) 84-2 p.141A Biophyscal publications
CrosRef
|
| 5 | Sequence information can be obtained from single DNA molecules
I. Braslavsky, B. Hebert, EP. Kartalov, SR. Quake,
Proc. Natl. Acad. Sci. U. S. A.
(2003) 100-7 p.3960 PNAS publications
CrosRef
|
| 6 | Microfluidic device reads up to four consecutive base pairs in DNA sequencing-by-synthesis
EP. Kartalov, SR. Quake,
Nucleic Acids Res.
(2004) 32-9 p.2873 PubMed CrosRef
|
.


